What is Downsampled Genes Data Analysis?
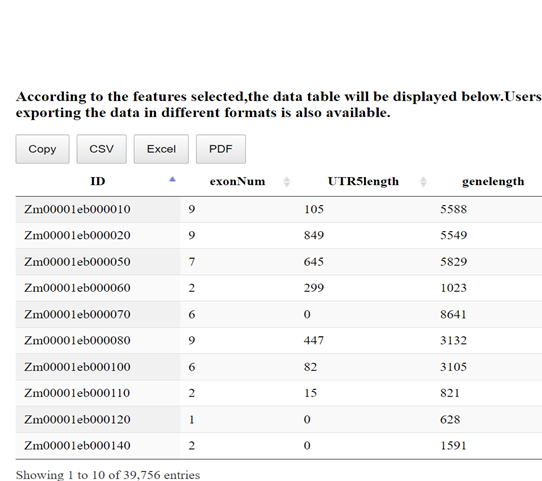
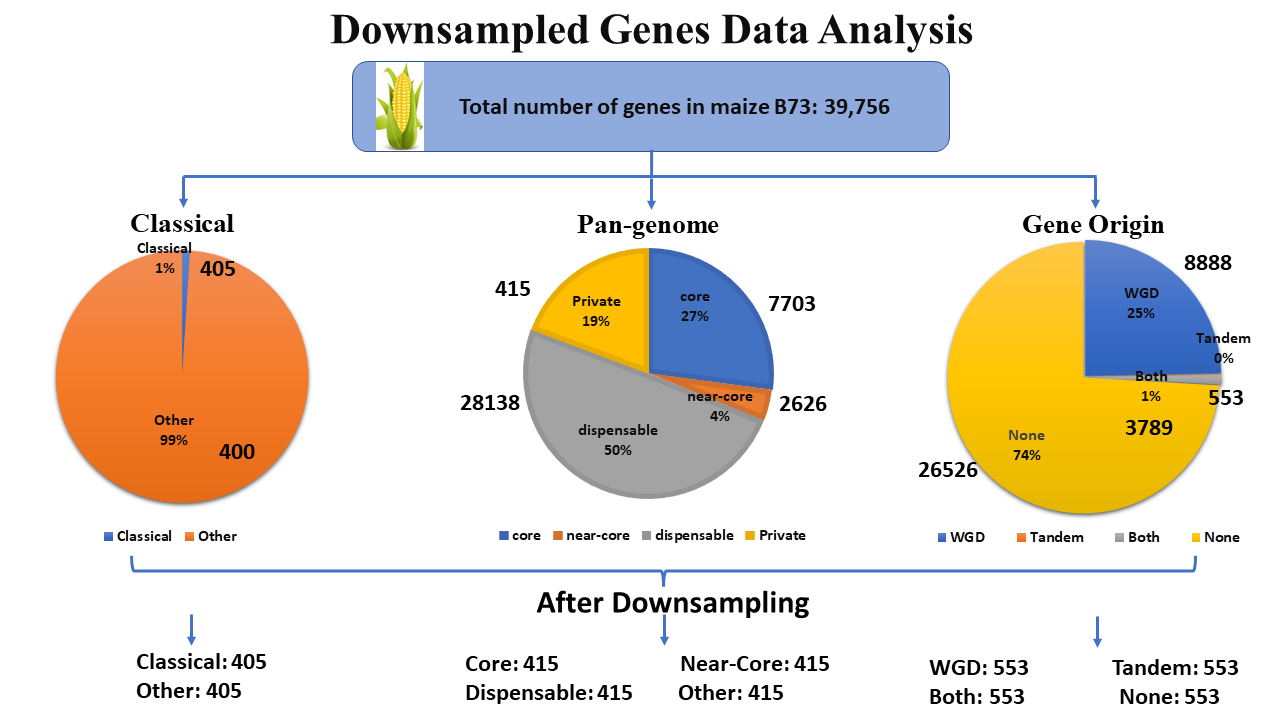
The analysis of downsampled genes data is crucial when dealing with imbalanced gene annotations in a reference genome. For example, in the maize pan-genome, 72% of genes are marked as core, while 28% are annotated as non-core. To address this imbalance, we employ the random downsampling method, allowing users to perform "Downsampled Analysis."
Random undersampling involves randomly removing samples from the majority class until a balanced class distribution is achieved. The Maize Feature Store (MFS) "Downsampled Genes Analysis" is designed to help users visualize diverse gene-based features for downsampled genes and explore relationships with gene annotations in categories such as Classical/Other, Pan-genome, and Gene Origin.
For more detailed information on using the "Downsampled Gene Analysis" module, please refer to the flow diagram or watch the video tutorial linked below.

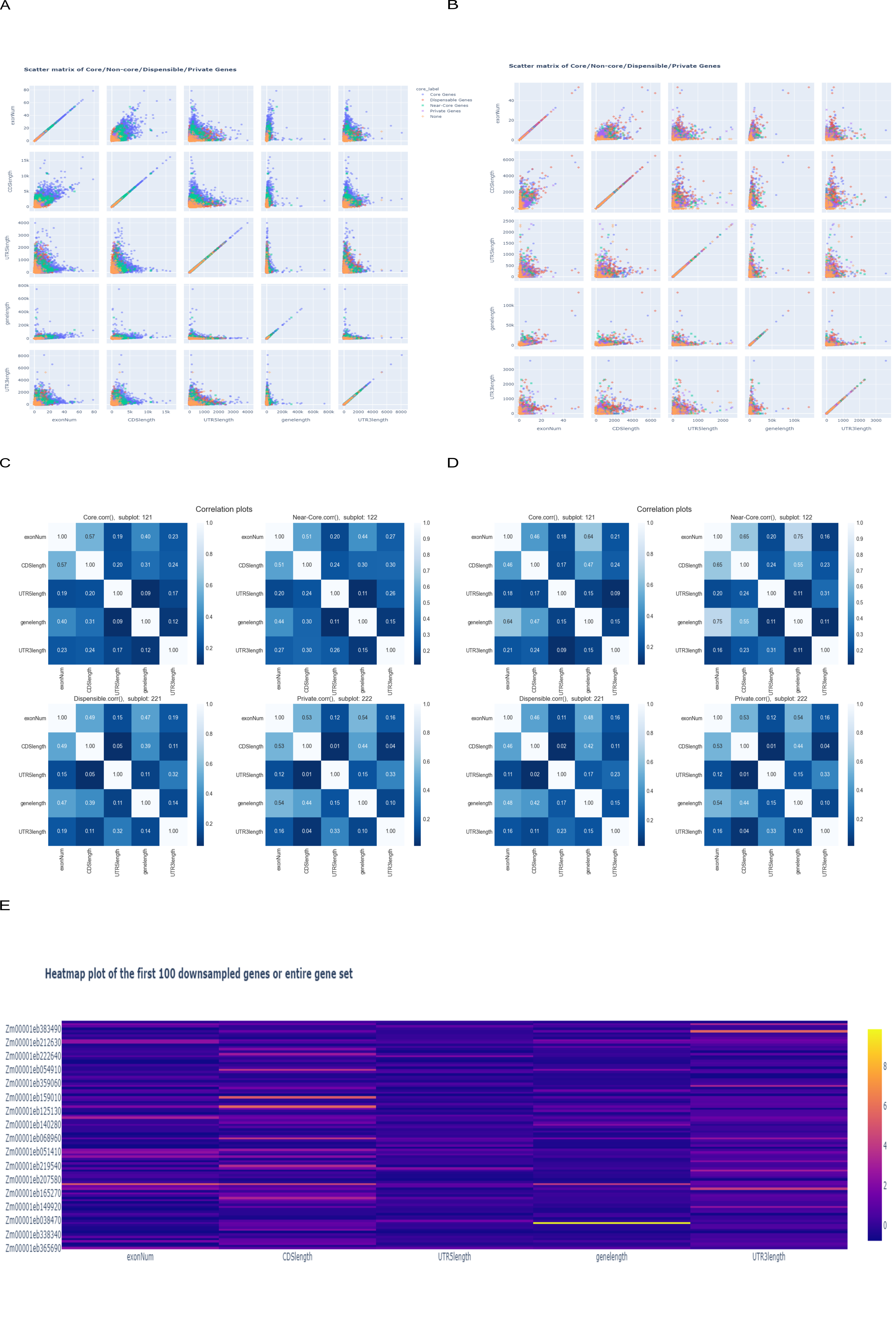
Omics datasets come in a diverse range, scale, and follow their own statistical distributions as they are collected from disparate sources, therefore data standardization becomes crucial for omics datasets. Outputs generated from non-standardized features are often skewed, deviated, and filled with outliers and anomalies. Thus, the advanced exploratory analysis such as Dendrograms, Hierarchical Heatmaps, Hierarchical Scatter plots, Heatmaps and PCA analysis demands high-level data preprocessing and normalization to balance out disproportionate weights across multiple variables. Data normalization transforms the multiscaled data all to the same scale, thereby improving the stability and performance of the learning algorithm. The Maize Feature Store application allows for the normalization of omics numerical features by centering the features with their mean and the standard deviation between 0 and 1 using the most common normalization method called Z-score normalization. In standardized z-score normalization, each feature is normalized as Z = ( X - X' ) / S, where X, X' and S are the feature, the mean and the standard deviation respectively.
- What is common among these genes in a given class?
- What are the relationships between: gene phenotype and gene length, copy number, expression levels and patterns, epigenetic markers, cross-species conservation, and SNP densities ?
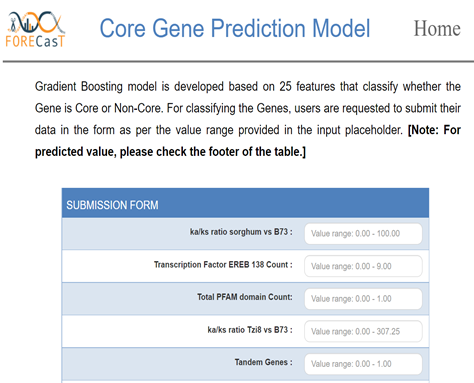
- Apply machine learning methods to predict important biological classifications.
- Provide tools to discover if genes within a class have distinct features and utilize it for constructing within- and cross-species prediction models.